Spike time data¶
Storing the times of spikes (action potentials) that are generated by neurons is one of the most common use cases.
The data shown below is simulation data created using a Leaky Integrate and Fire (LIF) model neuron. To run the example codes in this section you will need the lif model.
example code)¶ # create a new file overwriting any existing content
file_name = 'spike_tagging.nix'
file = nixio.File.open(file_name, nixio.FileMode.Overwrite)
# create a 'Block' that represents a grouping object. Here, the recording session.
# it gets a name and a type
block = file.create_block("block name", "nix.session")
# create a 'DataArray' to take the membrane voltage
data = block.create_data_array("membrane voltage", "nix.regular_sampled.time_series", data=voltage, label="membrane voltage", unit="mV")
# add descriptors for time axis
data.append_sampled_dimension(time[1]-time[0], label="time", unit="s")
# create the positions DataArray
positions = block.create_data_array("spike times", "nix.events.spike_times", data=spike_times)
positions.append_range_dimension_using_self()
# create a MultiTag
multi_tag = block.create_multi_tag("spike times", "nix.events.spike_times", positions)
multi_tag.references.append(data)
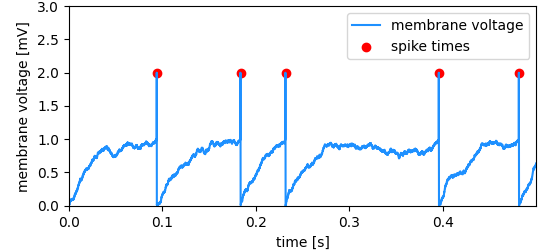
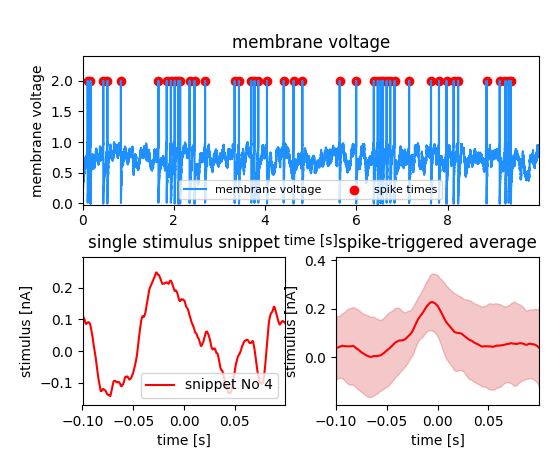
Spike times are marked in a neuron’s membrane potential (fake data, created with a LIF model neuron).
Adding features¶
The following code shows how to use the Features of the NIX-model. Suppose that we have the recording of a signal in which a set of events is detected. Each event may have certain characteristics one wants to store. These are stored as Features of the events. There are three different link-types between the features and the events stored in the tag. nix.LinkType.Untagged indicates that the whole data stored in the Feature applies to the points defined in the tag. nix.LinkType.Tagged on the other side implies that the position and extent have to be applied also to the data stored in the Feature. Finally, the nix.LinkType.Indexed indicates that there is one point (or slice) in the Feature data that is related to each position in the tag.
The following examples show how this works.
Tagging stimulus segments¶
Let’s say we record the neuronal activity and in a certain epoch of that recording a stimulus was presented. This time interval is annotated using a Tag. This inidicates the time in which the stimulus was on but we may also want to link the stimulus itself to it. The stimulus is also stored as a DataArray and can be linked to the Tag as an untagged Feature of it.
untaggedFeature.py.¶ stim = block.create_data_array("stimulus", "nix.sampled.time_series", data=stimulus,
label="current stimulus", unit="nA")
stim.append_sampled_dimension(stepsize, label="time", unit="s")
# create the Tag to highlight the stimulus-on segment
tag = block.create_tag("stimulus presentation", "nix.epoch.stimulus_presentation", [stim_onset])
tag.extent = [stim_duration]
tag.references.append(data)
# set stimulus as untagged feature of the tag
tag.create_feature(stim, nixio.LinkType.Untagged)
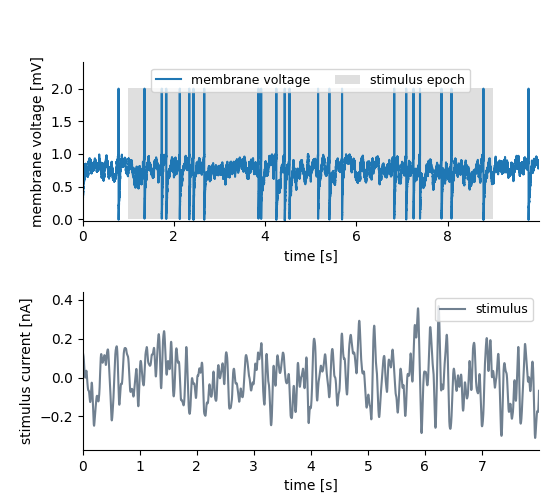
The recorded membrane voltage data is 10s long and we tag the interval between stimulus_onset and stimulus_onset + stimulus_duration (from 1 to 9 seconds). The stimulus itself is only 8s long and was played in the tagged interval. We use a Tag to bind stimulus and recorded signal together. The data stored in the “untagged” feature is the whole stimulus. The Tag’s position and extent do not apply to the stimulus trace.
Tagging with spike times¶
Tagged Features are used in cases in which the positions and extents of a tag also apply to another linked dataset. In the following example the spike times should also be applied to the stimulus that led to the responses. The stimulus is saved in an additional DataArray and is linked to the spike times as a Feature setting the LinkType to tagged.
taggedFeature.py.¶ data.append_sampled_dimension(stepsize, label="time", unit="s")
# create the positions DataArray
positions = block.create_data_array("spike times", "nix.events.spike_times", data=spike_times)
positions.append_range_dimension_using_self()
# create a MultiTag
multi_tag = block.create_multi_tag("spike times", "nix.events.spike_times", positions)
multi_tag.references.append(data)
# save stimulus snippets in a DataArray
stimulus_array = block.create_data_array("stimulus", "nix.sampled", data=stimulus, label="stimulus", unit="nA")
# add a descriptor for the time axis
stimulus_array.append_sampled_dimension(stepsize, label="time", unit="s")
# set stimulus as a tagged feature of the multi_tag
multi_tag.create_feature(stimulus_array, nixio.LinkType.Tagged)
# let's plot the data from the stored information
plot_data(multi_tag)
file.close()
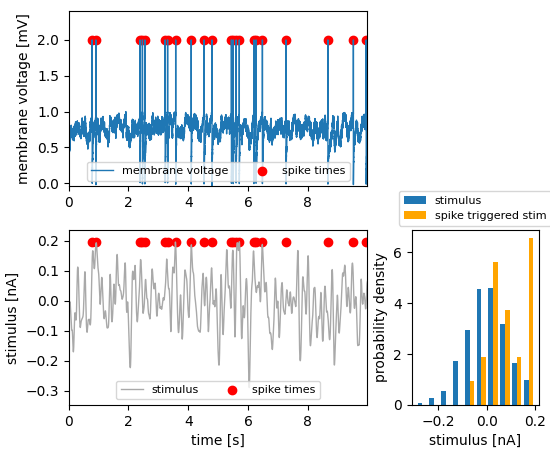
The spike times are used to tag the recording of the membrane voltage using a MultiTag. The stimulus is added to the MultiTag as a tagged feature. That is, the positions of the tag (the spike times) should be applied also to the stimulus. Extracting the feature_data gives the stimulus intensities at the times of the spikes, the orange distribution.
Storing wavelets as Features of the spike times¶
In the example, the signal is the membrane potential of a (model) neuron which was stimulated with some stimulus. The events are again the action potentials (or spikes) fired by that neuron. A typical analysis performed on such data is the Spike triggered average which represent the average stimulus that led to a spike. For each spike, a snippet of the respective stimulus is cut out and later averaged. In this example we store these stimulus snippets and link them to the events by adding a Feature to the MultiTag. There are three different flags that define how this link has to be interpreted. In this case there is one snippet for each spike. The index of each position has to be used as an index in the first dimension of the Feature data. The LinkType has to be set to indexed.
Indexed feature it must be organized that the first dimension represents the number of snippets. The second dimension is the time. (example code).¶ snippets = block.create_data_array("spike triggered stimulus", "nix.regular_sampled.multiple_series", data=sts, label="stimulus", unit="nA")
snippets.append_set_dimension()
snippets.append_sampled_dimension(stepsize, offset= -sta_offset * stepsize, label="time", unit="s")
# set snippets as an indexed feature of the multi_tag
multi_tag.create_feature(snippets, nixio.LinkType.Indexed)